EbbaBiolight NBIC Interview
Optotracing with EbbaBiolight
EbbaBiolight fluorescent tracer molecules are optotracers. Unlike conventional fluorescent dyes, optotracers bind promiscuously to a range of targets with repetitive motifs. EbbaBiolight has been shown to bind to curli and cellulose in Salmonella extracellular matrix [1,2], peptidoglycan and lipoteichoic acids in the cell envelope of Staphylococci [3], β-glucans from S. cerevisiae and Chitin in C. albicans [4]. Upon binding, the fluorescence intensity of the optotracer increases. This property makes it possible to use EbbaBiolight for live fluorescent tracking of microorganisms, without the need to wash away unbound molecules. It is possible to read out fluorescence intensity at the emission maximum (Emmax) when excited at or close to the excitation maximum (Exmax). This is useful for microscopy or fluorescence spectroscopy when straight-forward data analysis is required. Yet, due to the unique properties of the optotracers, a unique optical fingerprint is produced reflecting the specific nature of the target (sample composition) and environment (pH, osmolarity, polarity of the medium). This means that depending on the specific properties of the sample, Exmax or Emmax can shift, or the appearance of double peaks or shoulders might indicate binding to multiple targets. We therefore recommend acquiring fluorescence excitation and emission spectra whenever possible within experimental limitations. EbbaBiolight excitation- and emission spectra can be accessed here.
| Exmax | Emmax | Excitation spectrum (detect at Emmax) | Emission spectrum (excite at Exmax) | Recommended filter-sets |
|
|---|---|---|---|---|---|
| EbbaBiolight 480 | 420 nm | 480 nm | 300 - 450 nm | 450 - 800 nm | DAPI |
| EbbaBiolight 520 | 460 nm | 520 nm | 300 - 490 nm | 490 - 800 nm | FITC, GFP |
| EbbaBiolight 540 | 480 nm | 540 nm | 300 - 510 nm | 510 - 800 nm | FITC, GFP, YFP |
| EbbaBiolight 630 | 520 nm | 630 nm | 300 - 600 nm | 550 - 800 nm | PI, Cy3, TxRed, mCherry, Cy3.5 |
| EbbaBiolight 680 | 530 nm | 680 nm | 300 - 650 nm | 660 - 800 nm | PI, mCherry, Cy3.5 |
Read More:
- Choong FX et al. (2016) Real-Time optotracing of curli and cellulose in live Salmonella biofilms using luminescent oligothiophenes. npj Biofilms and Microbiomes, 2, 16024
- Choong FX et al. (2021) A semi high-throughput method for real-time monitoring of curli producing Salmonella biofilms on air-solid interfaces. Biofilm, 3, 100060
- Butina K. et al. (2020) Optotracing for selective fluorescence-based detection, visualization and quantification of live S. aureus in real-time. npj Biofilms and Microbiomes, 6(1), 35
- Kärkkäinen, E. et al. (2022) Optotracing for live selective fluorescence-based detection of Candida albicans biofilms. Frontiers in Cellular and Infection Microbiology, 12, 2235-2988
Antimicrobial peptides for targeting bacterial resistance
The problem of antibiotic resistance is leading to rising fatalities due to bacterial infections world wide and extensive research into antibiotic resistance has uncovered that more and more bacterial species are becoming immune to antibiotics. In order to protect against bacterial resistance, antimicrobial peptides are investigated for targeting bacterial resistance.
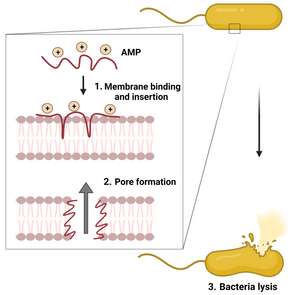
Antimicrobial peptides (AMPs) are natural cationic molecules that play an important role in the innate immune system of different organisms. AMPs differ from conventional antibiotics in terms of pharmacokinetics, a more narrow mutation window and fast effectiveness in killing bacteria as well as viruses and fungi. As AMPs have a natural ability to bind and permeate the negatively charged membranes of both Gram-positive and Gram-negative bacteria (see image), they offer a new and more varied approach to target infection. Moreover, the diversity offered in AMPs from their conserved nature has led to investigations into antiviral and antifungal solutions as well. Resistance to AMPs occurs much less than to conventional antibiotics.
The ability of bacteria to survive lethal concentrations of AMPs without a genetically encoded resistance mechanism has been studied by researchers at Freie Universität Berlin, in Germany who used two AMPs (mellitin and pexiganan) due to their broad applicability against Gram-positive and Gram-negative bacteria, as well as parasites and cancer cells. They have shown that as a response to sub-lethal doses of AMP’s, E.coli are able to develop not only a tolerance, but an increased persistence to lethal AMP treatment by producing curli in their biofilm. Their findings show that although AMPs offer a simple and effective solution to the antibiotic resistance crisis the world is facing today, they must also be used correctly. The risk of tolerance and resistance of bacteria to these AMP treatments exist and must be considered. EbbaBiolight can aid in establishing increased tolerance to AMPs due to its ability to detect and quantify curli content in the bacterial extracellular matrix of Gram-negative bacteria like E. coli or Salmonella.
EbbaBiolight fluorescence spectra
We named our EbbaBiolight molecules after their peak emission wavelength when they are bound to their target. That means, when EbbaBiolight is bound to a target, it will emit fluorescence at peak emission indicated by the number associated with its name.
To view the excitation and emission spectra, please select your EbbaBiolight below :