Monitoring curli in bacterial biofilms forming on semi-solid agar
This protocol describes how to use EbbaBiolight to visualise curli in biofilm forming on semi-solid agar in real-time. Curli is a functional amyloid produced by many Enterobactericeae involved in adhesion to surfaces, cell aggregation, and biofilm formation. EbbaBiolight are versatile molecules that have been reported to target various structures in the cell wall of gram-positive bacteria and the extracellular matrix of gram-negative bacteria. Curli has been identified as one of the major targets for EbbaBiolight in Salmonella biofilms using wildtype bacteria as well as curli deficient (ΔcsgA) strains. For reference, see Choong et al. (2021) Biofilms, 3, 100060. We recommend using fluorescent protein tagged bacteria to be able to distinguish between bacterial cells and curli in the bacterial extracellular matrix. The method described here is a semi-high throughput approach which allows for analysis at a faster pace and larger volume compared to conventional methods.
Materials:
Equipment:
Assay Procedure:
Note: When adapting this technique, please make sure to include relevant controls to verify that EbbaBiolight does not affect biofilm formation, to confirm curli as EbbaBiolight binding target and to exclude pH effects. Please be aware that fixation might alter the staining pattern of EbbaBiolight.
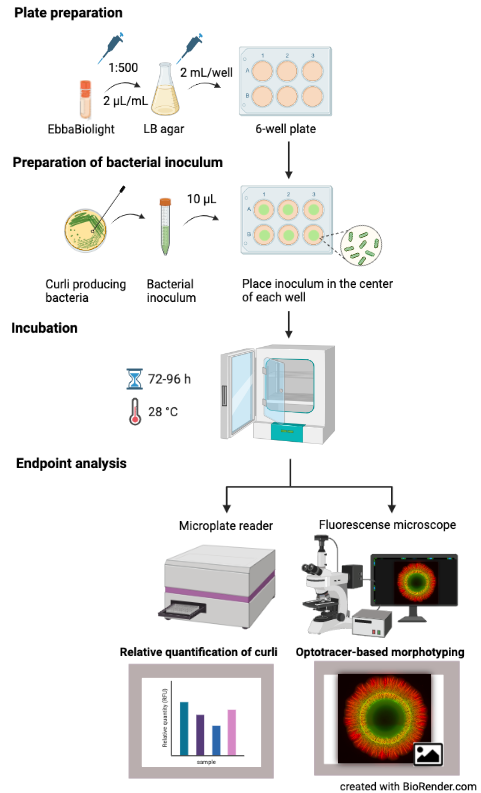
Materials:
- EbbaBiolight
- LB broth (w/o salt)
- LB agar (w/o salt)
- 6-well plate with cover
- Curli-producing bacteria on standard culture plate
Equipment:
- Incubator (28°C)
- Shaking Incubator (37°C)
- Microwave
- Fluorescence microscope OR Fluorescence Plate Reader
Assay Procedure:
- Prepare plates with EbbaBiolight supplemented LB agar:
- Microwave LB agar until it is melted.
- Let the microwaved agar rest at RT for 5-10 min.
- Add 2 µL/mL EbbaBiolight.
- Pour 2 mL of supplemented LB agar per well in a 6-well plate.
- Cover plates and let cool down at RT.
- Optional: Store at 4°C.
- Prepare bacterial inoculum:
- Pick a colony from a standard culture plate.
- Transfer colony to LB broth.
- Prepare an overnight or exponential culture under continuous shaking at 37°C.
- Transfer 10 µl of bacterial inoculum to the centre of a dedicated well on the previously prepared 6-well plate.
- Incubation:
- incubate the plate in an incubator at 28°C for as long as required for the biofilm to form.
- Endpoint analysis:
- Analyse in fluorescent microscope for optotracer based morphotyping OR in Fluorescence Microplate Reader for curli quantification.
Optotracing with EbbaBiolight
EbbaBiolight fluorescent tracer molecules are optotracers. Unlike conventional fluorescent dyes, optotracers bind promiscuously to a range of targets with repetitive motifs. EbbaBiolight has been shown to bind to curli and cellulose in Salmonella extracellular matrix [1,2], peptidoglycan and lipoteichoic acids in the cell envelope of Staphylococci [3], β-glucans from S. cerevisiae and Chitin in C. albicans [4]. Upon binding, the fluorescence intensity of the optotracer increases. This property makes it possible to use EbbaBiolight for live fluorescent tracking of microorganisms, without the need to wash away unbound molecules. It is possible to read out fluorescence intensity at the emission maximum (Emmax) when excited at or close to the excitation maximum (Exmax). This is useful for microscopy or fluorescence spectroscopy when straight-forward data analysis is required. Yet, due to the unique properties of the optotracers, a unique optical fingerprint is produced reflecting the specific nature of the target (sample composition) and environment (pH, osmolarity, polarity of the medium). This means that depending on the specific properties of the sample, Exmax or Emmax can shift, or the appearance of double peaks or shoulders might indicate binding to multiple targets. We therefore recommend acquiring fluorescence excitation and emission spectra whenever possible within experimental limitations. EbbaBiolight excitation- and emission spectra can be accessed here.
| Exmax | Emmax | Excitation spectrum (detect at Emmax) | Emission spectrum (excite at Exmax) | Recommended filter-sets |
|
|---|---|---|---|---|---|
| EbbaBiolight 480 | 420 nm | 480 nm | 300 - 450 nm | 450 - 800 nm | DAPI |
| EbbaBiolight 520 | 460 nm | 520 nm | 300 - 490 nm | 490 - 800 nm | FITC, GFP |
| EbbaBiolight 540 | 480 nm | 540 nm | 300 - 510 nm | 510 - 800 nm | FITC, GFP, YFP |
| EbbaBiolight 630 | 520 nm | 630 nm | 300 - 600 nm | 550 - 800 nm | PI, Cy3, TxRed, mCherry, Cy3.5 |
| EbbaBiolight 680 | 530 nm | 680 nm | 300 - 650 nm | 660 - 800 nm | PI, mCherry, Cy3.5 |
Read More:
- Choong FX et al. (2016) Real-Time optotracing of curli and cellulose in live Salmonella biofilms using luminescent oligothiophenes. npj Biofilms and Microbiomes, 2, 16024
- Choong FX et al. (2021) A semi high-throughput method for real-time monitoring of curli producing Salmonella biofilms on air-solid interfaces. Biofilm, 3, 100060
- Butina K. et al. (2020) Optotracing for selective fluorescence-based detection, visualization and quantification of live S. aureus in real-time. npj Biofilms and Microbiomes, 6(1), 35
- Kärkkäinen, E. et al. (2022) Optotracing for live selective fluorescence-based detection of Candida albicans biofilms. Frontiers in Cellular and Infection Microbiology, 12, 2235-2988
