Antibiotic Susceptibility of Salmonella using EbbaBiolight supplemented agar
Antibiotic susceptibility testing is widely performed in clinical microbiology labs. The disk diffusion method is the gold standard for confirming the susceptibility of bacteria. Using this method, introduced by Bauer and Kirby in 1956, a standardized bacterial suspension is prepared and inoculated onto solidified agar and an antibiotic-treated paper (disk) is tapped on the inoculated plate. The disc containing the antibiotic is allowed to diffuse through the solidified agar, resulting in the formation of an inhibition zone after the overnight incubation at 35 °C. The size of the inhibition zone formed around the paper disc is measured and the assay generally takes 16–24 h. Assessing the inhibition zone using this assay gives a relatively clear picture of the antibiotic susceptibility of the bacteria, yet it is unclear if the antibiotic also impacts the extracellular matrix in which the bacterial cells are embedded. As a result, patients may receive an insufficient antibiotic dose and treatment, putting them at risk of recurrent infections. Furthermore, inappropriate antibiotic use contributes to the development of antimicrobial resistance. In a recent study, published in the Journal Biofilm, researchers at AIMES located at Karolinska Institutet in Stockholm, Sweden, employed an EbbaBiolight-based biofilm assay to shed light on the interaction between antibiotics and the extracellular matrix formed by Salmonella colonies on agar plates. The agar was supplemented with EbbaBiolight 680 and, in accordance with the disk-diffusion assay, antibiotic discs were used to assess the antibiofilm efficiency of ten clinically relevant antibiotics for Salmonella. While the majority of antibiotics demonstrated the ability to decrease colony growth, only 4 antibiotics exhibited the capacity to inhibit the formation of the curli-based extracellular matrix. Notably, colonies lacking curli were more susceptible to antibiotics than colonies with intact curli synthesis. This suggests that antibiotics that can reduce curli production have the potential to enhance the sensitivity of colonies to other antibiotics, making them valuable candidates for inclusion in combination therapies for more effective treatment of biofilm infections. This study emphasizes the need for biofilm-specific treatment strategies and readily available tools for their development. In this context, the EbbaBiolight molecules can provide an easy to use, readily available tool for studying biofilm formation and evaluating the effects of antimicrobials.
Read More:
Uncovering bacteria & funghi interactions
Microbial populations communicate to achieve tasks that a single organism can not. Therefore, understanding the intricate interactions between different microorganisms is an essential missing piece of scientific knowledge. Elucidating the details of the interaction between Pseudomonas aeruginosa and Candida albicans is of particular interest for biochemical science and clinical practice as these pathogens are associated with a high rate of nosocomial infections worldwide. It is known that Pseudomonas and Candida are commonly found together and Pseudomonas is known to reduce the growth of the fungus both via physical contact and the release of chemical signals. However, conventional methods for studying polymicrobial interactions often encounter limitations in precisely discerning specific interactions between microorganisms. In a recent study, researchers from the group of Xiaolong Luo at the Catholic University of America developed a microfluidics platform to try and address this issue. Their innovative approach allowed them to study the complex interplay between Pseudomonas aeruginosa and Candida albicans by growing the bacteria and the fungus in side-by-side microchannels separated by a thin membrane with anti-microbial properties. This approach not only lets the scientists prevent the two species from physically interacting, but also facilitates the continuous monitoring and quantitative analysis of their mutual interactions. Using their system, the researchers were able to confirm that “chemical” contact with Pseudomonas reduces the growth rate of Candida and pushes it towards its hyphal form. However, they also observed how the contact with Candida albicans seems to enhance Pseudomonas growth. At this point, EbbaBiolight 480 was used to confirm that this increased growth is due to the formation of bigger, faster-growing biofilms. Their unique setup and the possibility of using Ebbabiolight 480 in live cultures allowed Pham and colleagues to observe the interaction between Pseudomonas and fully formed Candida albicans hyphae: after 15 hours of co-culture, Pseudomonas busts through the anti-microbial membrane and starts invading the microchannel in which Candida had been growing. In this invasion, the bacteria uses the hyphae as a walkway through the hydrogel matrix, and grows around the fungus, rather than in circular colonies as it normally does. The work from Pham and their colleagues highlights the significance of microbial interactions in invasive infections, as well as the need of better tools to observe these interactions, as the conventional methods could easily be misleading: without the ability to physically separate the two cultures and tracking live the formation of biofilm, observing the way in which Pseudomonas biofilm grows around Candida’s hyphae would not have been possible.
Read More:
Uncovering bacteria & funghi interactions
Microbial populations communicate to achieve tasks that a single organism can not. Therefore, understanding the intricate interactions between different microorganisms is an essential missing piece of scientific knowledge. Elucidating the details of the interaction between Pseudomonas aeruginosa and Candida albicans is of particular interest for biochemical science and clinical practice as these pathogens are associated with a high rate of nosocomial infections worldwide. It is known that Pseudomonas and Candida are commonly found together and Pseudomonas is known to reduce the growth of the fungus both via physical contact and the release of chemical signals. However, conventional methods for studying polymicrobial interactions often encounter limitations in precisely discerning specific interactions between microorganisms.
In a recent study, researchers from the group of Xiaolong Luo at the Catholic University of America developed a microfluidics platform to try and address this issue. Their innovative approach allowed them to study the complex interplay between Pseudomonas aeruginosa and Candida albicans by growing the bacteria and the fungus in side-by-side microchannels separated by a thin membrane with anti-microbial properties. This approach not only lets the scientists prevent the two species from physically interacting, but also facilitates the continuous monitoring and quantitative analysis of their mutual interactions.
Using their system, the researchers were able to confirm that “chemical” contact with Pseudomonas reduces the growth rate of Candida and pushes it towards its hyphal form. However, they also observed how the contact with Candida albicans seems to enhance Pseudomonas growth. At this point, EbbaBiolight 480 was used to confirm that this increased growth is due to the formation of bigger, faster-growing biofilms.
Their unique setup and the possibility of using Ebbabiolight 480 in live cultures allowed Pham and colleagues to observe the interaction between Pseudomonas and fully formed Candida albicans hyphae: after 15 hours of co-culture, Pseudomonas busts through the anti-microbial membrane and starts invading the microchannel in which Candida had been growing. In this invasion, the bacteria uses the hyphae as a walkway through the hydrogel matrix, and grows around the fungus, rather than in circular colonies as it normally does.
The work from Pham and their colleagues highlights the significance of microbial interactions in invasive infections, as well as the need of better tools to observe these interactions, as the conventional methods could easily be misleading: without the ability to physically separate the two cultures and tracking live the formation of biofilm, observing the way in which Pseudomonas biofilm grows around Candida’s hyphae would not have been possible.
Tracking of biofilm formation in Burkholderia
Bacteria and fungi produce biofilms whenever they adhere to a surface to protect themselves from environmental stressors, including antibiotics. Biofilm-related infections are therefore often hard to treat and do not respond well to antibiotic treatment. As an opportunistic pathogen of the respiratory tract Burkholderia cenocepacia is often responsible for hospital-acquired infections. While Burkholderia has an innate resistance to antibiotics, it also forms biofilms, rendering the treatment of these infections especially cumbersome. Burkholderia shares several similarities with Pseudomonas aeruginosa, both in the type of infections it causes, and the difficulty of treatment. In Pseudomonas, the wsp (wrinkly spreader phenotype) system is a chemosensory signal transduction system that triggers biofilm formation. The system is also present in Burkholderia. So, Dr. Andrea Sass and their colleagues at Ghent University wondered if the wsp cluster of genes serves a similar purpose in both species. In Burkholderia, the wsp cluster is regulated by methylation in the promoter of an upstream gene called wspH. Deletion of the enzyme that methylates the promoter increases expression of both wspH and the wsp cluster downstream of it, suggesting that these genes are part of the same operon. However, wspH is fairly far from the rest of the cluster, and thus its role in the regulation of the wsp cluster has not been investigated before. With a clever genetic approach, the researchers could indeed determine that wspH is expressed together with the other wsp genes in Burkholderia. After figuring out the components of the wsp cluster, the researchers wanted to determine the effects of the expression of this operon in the formation of biofilms. They used EbbaBiolight 680 to track the formation of a pellicle biofilm at the air-liquid interface in static cultures of Burkholderia, and found that mutants overexpressing wspH; were much faster in forming a biofilm than others. This was accompanied by an increase of c-GMP within the bacterial cells, which is a common regulator of cell motility and biofilm formation. The c-GMP signal resulted in the expression of the wsp cluster, which produces exopolysaccharides that increase the hydrophobic properties of the cell surface. Overexpressing only the other wsp genes - and not wspH - had, instead, only mild effects on the formation of biofilms, indicating a central role of wspHin the initiation of biofilm formation.
Being able to observe the temporal dynamics of biofilm formation and the effects of putative regulatory factors in live cultures is key to the development of new treatments. These treatments may combat the microbe-protective biofilms and prevent re-infections, and non-toxic trackers like EbbaBiolight are an invaluable tool to achieve this.
Read More:
Tracking fungal biofilm formation
Candida albicans is a commensal fungus that lives among the gut flora of 40 to 60% of healthy individuals, where it presents as ovoid “yeast” cells. Changes in its environment can trigger a switch towards the formation of multicellular hyphae which is thought to be a crucial step for the virulence of the fungus as intertwined hyphal filaments enable the formation of a biofilm. This means that, through biofilm formation, C. albicans can become an opportunistic pathogen and cause infections, which is particularly problematic in immunocompromised patients.
When forming a biofilm, the fungus surrounds itself with “fungal superglue” - an extracellular matrix (ECM) of β-1,3-glucans, lipids, and nucleic acids - that protects it not only against the immune system but also from antifungal drugs. This makes treatment of C. albicans infections particularly challenging with mortality rates of up to 40% in cases of systemic candidiasis. The limited number of detection methods for C. albicans further emphasises the need for technical development in this area. While biofilms can be measured by biomass or metabolic activity, distinguishing between growth states is hard, although critical for identifying active disease.
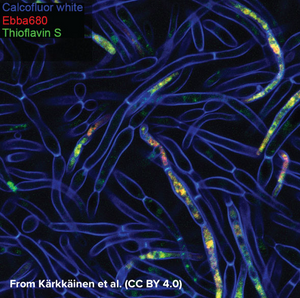
Kärkkäinen et al. from AIMES at Karolinska Institutet in Sweden took on the challenge to try to identify specific markers for the biofilm status using fluorescent probes. The researchers used a range of fluorescent probes for detailed analysis of yeast cells and hyphae. Fluorescently tagged Concavalin A which is a Carbohydrate-Binding-Protein and Calcofluor white which binds to cellulose and chitin were used to label the cell wall. DAPI was used as a nuclear stain, MDY-64 was used to label cell membrane and vacuoles and Thioflavin S was used to label densely packed and dispersed amyloids. Whereas the use of these labels gave a very detailed view on the anatomy of cellular yeast and yeast hyphae in biofilms, no clear cues for differentiation of yeast cells and biofilm was found that didn’t involve microscopic investigation. As common markers failed to differentiate yeast cells from biofilms, the researchers tested if EbbaBiolight 680 can be used to discern between cell wall polysaccharides and thus differentiate between cellular yeast and biofilm. Initially, the researchers tested if EbbaBiolight 680 binds to purified components of the yeast cell wall and found that EbbaBiolight 680 is unable to detect mannans, but binds to β-glucans and chitin. Moreover, the researchers found that the emission spectrum of EbbaBiolight 680 bound to β-glucans differs from the emission spectrum of EbbaBiolight 680 bound to chitin, suggesting that EbbaBiolight 680 might be used to distinguish between the two.
Therefore, the researchers used EbbaBiolight 680 to label live-cultures of C. albicans as a next step. Interestingly, they found that the signal from the cell walls was relatively weak compared to the bright signal obtained from the intracellular amyloids. Furthermore, significant differences were found in the emission spectra of EbbaBiolight 680 bound to intracellular amyloid structures in yeast cells compared to cells growing in a biofilm.. EbbaBiolight 680 bound to amyloids in yeast cells showed a broad emission spectrum with two peaks; one at 498 nm and one at 614 nm whereas EbbaBiolight 680 bound to amyloids in yeast biofilm showed an emission spectrum with one peak at 634 nm. Thus, the ratio of the fluorescence intensity (RFU) at 498 nm and at 634 nm was used as a measure to tell the difference between cellular yeast and yeast biofilm.
In summary, the publication by Kärkkäinen et al. shows the potential for EbbaBiolight to differentiate between yeast cells and yeast biofilm and thereby solve a clinical problem providing means of differentiating between the commensal and pathogenic lifestyle of a microorganism.
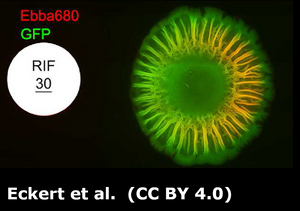
Read More:
Watching bacterial cities grow
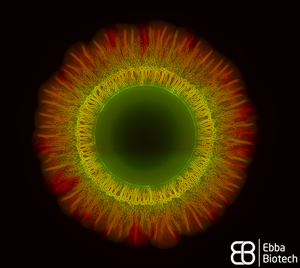
When thinking about bacteria, we often imagine bacterial cells floating in liquid medium. But, in reality, this planktonic lifestyle is only a very small episode in the bacterial life cycle. To be able to survive environmental stress like nutrient shortages and attacks launched by cleaning brushes, antibiotics or disinfectants, they attach to surfaces and build colonies that are reinforced with a self-produced extracellular matrix (ECM). The ECM acts as glue to keep the colony together so that the single bacteria can support each other but also act as a protective barrier. The conglomerate of bacteria in their self-produced extracellular matrix is called biofilm. Researchers from the Centre for the Advancement of Integrated Medical and Engineering Sciences (AIMES) at Karolinska Institutet in Sweden succeeded to take a unique look at Salmonella bacteria growing biofilm on semi-solid agar substrate. To be able to do that, the researchers used Salmonella that were previously tagged with green fluorescent protein (GFP) and added EbbaBiolight 680 to the semi-solid agar substrate on which the bacterial colonies were growing. They presented their findings in a study by Choong et al., which was recently published in the scientific journal Biofilms. EbbaBiolight are small, non-toxic fluorescent tracer molecules (optotracers) that don't disturb bacterial growth or biofilm formation. Also, EbbaBiolight fluorescence is switched-on only when the optotracer is bound to its target, which in the case of EbbaBiolight 680 has been identified as the ECM protein curli. Thus, EbbaBiolight 680 labels the ECM of gram-negative, curli producing bacteria like Salmonella or E. coli and allows imaging with extremely low background fluorescence. To be able to monitor many cultures simultaneously in an automated fashion, the researchers devised a semi-high throughput approach in which they used 6-well plates containing agar that was previously supplemented with EbbaBiolight 680 and placed a small amount of bacteria right in the middle of each dish to start-off the colony growth and biofilm formation.
The properties of the EbbaBiolight molecule allowed the researchers to monitor ECM formation in great detail and together with GFP-expressing bacteria revealed specific regions of active bacterial growth and the spatiotemporal co-localisation of biofilm components that expanded in a well-structured manner, projecting radial channel patterns (see video). Using GFP and EbbaBiolight 680 as fluorescent markers for bacteria and ECM does not only support fluorescence imaging but also enables fluorescence spectroscopy which facilitates data collection and subsequent analysis of the growth pattern in different regions of the bacterial biofilm. In summary, the semi-high throughput methodology using EbbaBiolight 680 provides a dynamic real-time analysis of biofilm formation on solid surfaces and might advance biofilm research by allowing the researchers to take biofilms and their structure into consideration when studying bacterial infections.
Read More:
Are antimicrobial peptides ending the Antibiotic Crisis?
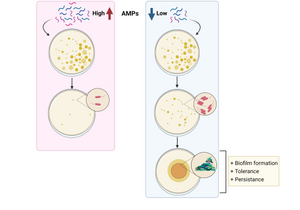
Antibiotic resistance is a major worldwide threat, rising to dangerous levels in which first-line antibiotics will no longer be effective. In this study researchers at Freie Universität Berlin, Germany described the use of antimicrobial peptides (AMPs) as an alternative approach against bacterial resistance. Using sub-lethal doses of AMPs, they demonstrated that bacteria develop both tolerance and persistence to the treatment by increasing the production of curli and colonic acid, which are extracellular matrix (ECM) components important for biofilm formation. The authors studied the emergence of priming and resistance to AMPs by priming Escherichia coli with sub-lethal doses of mellitin and pexiganan. They used EbbaBiolight 680 to label the extracellular matrix component curli and found increased curli production in bacteria primed with mellitin. Together with findings from transcriptomics and killing assays, the authors were able to conclude that priming with sublethal doses of AMPs, leads to an increase in tolerance and persistence, promoting bacteria survival compared to their native counterparts.
Altogether, the study shows that exposure to sub-lethal doses of AMPs correlates with an increase in tolerance and persistence by promoting biofilm formation, and hence, bacterial survival and colonisation. The results suggest that for a correct use of AMPs, a fast increase in their concentration should be provided to avoid bacterial priming, which will translate into resistance evolution. The findings shed light on how non-inherited resistance is acquired and open new paths to investigate measures to combat bacterial resistance.
Image: Graphical summary of the study. While high concentration of AMPs avoids bacterial survival, low concentration results in increased biofilm formation, tolerance and persistence, correlated with evolution to resistance.
Note: In the publication by Rodriguex-Rojas et al., the previous commercial name for EbbaBiolight is used. All claims made for ECtracer 680 in this publication can be applied to EbbaBiolight 680.
Read More:
EbbaBiolight-like molecule for detection and quantification of bacteria
Fast and reliable testing for pathogenic bacteria like Stapylococcus aureus (S. aureus) is highly desirable in the clinic as it can cause deep-seated infections such as osteomyelitis and endocarditis and is the major cause for hospital acquired infections of surgical wounds or indwelling medical devices. S. aureus is known for its ability to become resistant to antibiotics with the Methicillin-resistant S. aureus (MRSA) as the most prominent example causing epidemic waves of hospital-acquired and community-associated infections. To prevent the spread of resistant strains of S. aureus, rapid and widespread testing of patients and hospital environments is of utter importance.
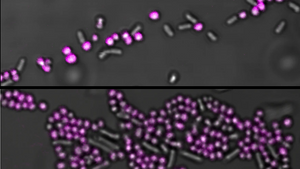
Researchers at the Karolinska Institute in Sweden have used an EbbaBiolight-like Molecule as fluorescent label for Staphylococci in mixed liquid cultures. Added to a mixed species sample containing Staphylococcus aureus and Salmonella Enteritidis, Staphylococcus epidermidis and Escherichia coli or Staphylococcus aureus and Enterococcus faecalis, Staphylococci are immediately distinguished by a bright fluorescent label.
As background fluorescence is almost non-existent using the EbbaBiolight-like Molecule, washing steps are obsolete and, importantly, quantitative studies of live, growing bacteria are feasible as the number of bacteria linearly correlates with the fluorescence intensity. This feature was used in a high throughput approach where a transposon mutagenesis library with >2000 strains was used to determine the binding target of the EbbaBiolight-like Molecule by tracking bacterial growth by means of optical density (OD) measurement and by analyzing binding of the EbbaBiolight-like Molecule by detecting relative fluorescence intensity.
The results from the high-throughput assay indicated that the EbbaBiolight-like Molecule likely binds to cell envelope structures. Further analysis confirmed that cell wall components lipoteichoic acid (LTA), peptidoglycan (PGN) as well as wall teichoic acids (WTA) act as epitopes for the EbbaBiolight-like Molecule. Super-resolution microscopy using an Airyscan detector was used to confirm localization of the EbbaBiolight-like Molecule in the cell wall including the division septum of Staphylococcus epidermidis.
Interestingly, selective binding of S. aureus was achieved, even when mixed with E. faecalis which also carries an exposed cell envelope. This was explained by hydrophobic interactions playing a major role in binding of the EbbaBiologht-like. At pH5, when cell envelope components are protonated and electrostatic repulsion is reduced the EbbaBiolight-like Molecule indeed binds to gram-positive S. aureus and E. faecalis but not gram-negative S. Enteritidis.
Read More:
EbbaBiolight-like molecule detects biomarker for recurrent infection
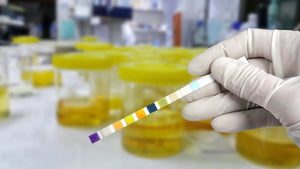
Bacteria such as Escherichia coli (E. coli) are known to hide during an infection by encasing themselves in extracellular matrix containing amyloid proteins and cellulose. When they grow like this, clusters of bacteria are called biofilm and they are hard to detect and partly resistant to antibiotics. The same bacteria are known for causing urinary tract infections (UTIs), which often turn out to be a recurrent illness. UTIs are very common and about 50 % of women have had such an infection at some point in their life. Uncomplicated cases of UTIs are treated with antibiotics, but in case of chronic and recurrent UTI, antibiotic resistance is an important problem. If antibiotics cannot combat the bacteria, kidney infections or even sepsis can occur and cause serious complications. Researchers at Karolinska Institutet in Sweden asked the question if the reason for chronic and recurrent UTI might be related to the extracellular matrix surrounding E. coli, rather than the bacteria themselves. Therefore, they used an EbbaBiolight-like Molecule to detect cellulose, which humans do not produce, but which is an abundant component in E. coli biofilm. So, if cellulose is detected in a human urine sample, it is likely to be of bacterial origin. The researchers collected urine samples from Karolinska University Hospital and tested the urin for presence of cellulose using a rapid and simple method. Using an EbbaBiolight-like Molecule, an assay was developed which allowed biofilm screening of urine samples within 45 min and it was shown that about 15 % of urine samples contained cellulose and were therefore related to biofilm forming E. coli. Linking biofilm-related infections to recurrent UTIs is an important step for future diagnostic efforts as this information can help to determine the correct course of treatment and thereby help to avoid serious complications that can be harmful to patients.
Read More:
EbbaBiolight-like molecules reveals Salmonella biofilm secrets
A 2016 study by the Swedish Medical Nanoscience Center at Karolinska Institutet used an EbbaBiolight-like Molecule to detect growth of Salmonella biofilm. This unique fluorescent tracer molecule is non-bactericidal and is therefore capable to dynamically follow the formation of curli fibres and cellulose in Salmonella enterica biofilm in real time. In the scientific study, wild-type Salmonella enterica serovar Enteritidis (S. Enteritidis) producing both cellulose and curli in its extracellular matrix (ECM) was compared to isogenic mutants of S. Enteritidis lacking cellulose, curli or both ECM components. Visualization of the ECM components curli and cellulose during growth in liquid medium is not possible with standard methods. Furthermore, the commonly used rdar morphotyping technique on agar plates requires at least 3 days of incubation and use of toxic chemicals. In contrast, by adding the EbbaBiolight-like fluorescent tracer molecule to the liquid culture, it becomes possible to track the production of cellulose and curli while the culture is still undergoing exponential growth and later entering the stationary phase (0-48 h). On agar, biofilms containing cellulose and/or curli were identified already after 24 h of incubation.
The EbbaBiolight-like molecule used in this study is a fluorescent tracer molecule that becomes highly fluorescent when binding to its target. Therefore, no washing is required in this procedure. The fluorescence spectrum is acquired using a plate reader or a fluorescence microscope. An increase in fluorescence and a shift in the maximum emission wavelength is used to indicate the presence of cellulose, curli or both ECM components in the biofilm. The researchers demonstrated that the molecule can be used to study biofilm formation in liquid cultures and on agar plates. They also showed applications to visualize surface-bound biofilm formation at the air-liquid interface, intracellular biofilm in macrophages in a cell culture infection experiment and even in infected liver tissue. This shows the versatility of EbbaBiolight for biofilm identification and visualization in liquid, cell culture infection experiments and even in infected tissues. Due to its unique properties, EbbaBiolight can be used to not only identify, but also to track biofilm formation and composition in various conditions. This marks EbbaBiolight as an excellent tool to investigate biofilm composition and dynamics as it has not been possible before.