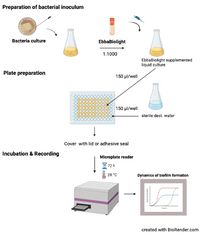
This protocol describes how to monitor kinetics of Salmonella extracellular matrix (curli) production in liquid culture. The method described here is based on Choong et al. (2016) npj Biofilms and Microbiomes, 2, 16024 where isogenic mutants of S. Enteritidis were used to identify the extracellular matrix components curli and cellulose as targets for optotracer binding. When used as described, EbbaBiolight does not label Salmonella cell wall and does not influence biofilm formation.
Materials:
- EbbaBiolight
- Growth medium
- Bacteria on standard culture plate
- 96-well plate (round bottom) with cover
- Deionized water
Equipment:
- Incubator (28°C)
- Shaking Incubator (37°C)
- Fluorescence plate reader
Assay Procedure:
- Prepare bacterial inoculum:
- Pick a colony from a standard culture plate.
- Transfer colony to growth medium.
- Prepare an overnight or exponential culture under continuous shaking at 37°C.
- Dilute bacterial culture 1:100 in fresh growth medium.
- Add EbbaBiolight (1:1000) and mix gently.
- Prepare 96-well plate:
- Use 100-200 µl liquid culture supplemented with EbbaBiolight per well.
- Fill unused wells with sterile water to avoid drying during incubation.
- Seal the plate with a cover.
- Incubation & Readout:
- incubate the 96-well plate directly in the plate reader under static conditions.
- Record excitation- and emission at regular time points during biofilm growth.
Read more →
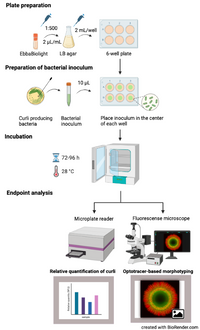
This protocol describes how to use EbbaBiolight to visualise curli in biofilm forming on semi-solid agar in real-time. Curli is a functional amyloid produced by many Enterobactericeae involved in adhesion to surfaces, cell aggregation, and biofilm formation. EbbaBiolight are versatile molecules that have been reported to target various structures in the cell wall of gram-positive bacteria and the extracellular matrix of gram-negative bacteria. Curli has been identified as one of the major targets for EbbaBiolight in Salmonella biofilms using wildtype bacteria as well as curli deficient (ΔcsgA) strains. For reference, see Choong et al. (2021) Biofilms, 3, 100060. We recommend using fluorescent protein tagged bacteria to be able to distinguish between bacterial cells and curli in the bacterial extracellular matrix. The method described here is a semi-high throughput approach which allows for analysis at a faster pace and larger volume compared to conventional methods.
Materials:
- EbbaBiolight
- LB broth (w/o salt)
- LB agar (w/o salt)
- 6-well plate with cover
- Curli-producing bacteria on standard culture plate
Equipment:
- Incubator (28°C)
- Shaking Incubator (37°C)
- Microwave
- Fluorescence microscope OR Fluorescence Plate Reader
Assay Procedure:
- Prepare plates with EbbaBiolight supplemented LB agar:
- Microwave LB agar until it is melted.
- Let the microwaved agar rest at RT for 5-10 min.
- Add 2 µL/mL EbbaBiolight.
- Pour 2 mL of supplemented LB agar per well in a 6-well plate.
- Cover plates and let cool down at RT.
- Optional: Store at 4°C.
- Prepare bacterial inoculum:
- Pick a colony from a standard culture plate.
- Transfer colony to LB broth.
- Prepare an overnight or exponential culture under continuous shaking at 37°C.
- Transfer 10 µl of bacterial inoculum to the centre of a dedicated well on the previously prepared 6-well plate.
- Incubation:
- incubate the plate in an incubator at 28°C for as long as required for the biofilm to form.
- Endpoint analysis:
- Analyse in fluorescent microscope for optotracer based morphotyping OR in Fluorescence Microplate Reader for curli quantification.
Read more →
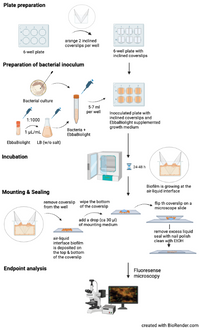
This protocol describes how to grow Salmonella biofilm at an air-liquid interface using inclined glass coverslips and how to visualize Salmonella extracellular matrix component curli using EbbaBiolight. The method described here is based on Choong et al. (2016) npj Biofilms and Microbiomes, 2, 16024 where isogenic mutants of S. Enteritidis were used to identify the extracellular matrix components curli and cellulose as targets for optotracer binding. When used as recommended, EbbaBiolight does not label Salmonella cell wall and does not influence biofilm formation. If adding EbbaBiolight during biofilm growth is not feasible, it can also be applied after the biofilm has assembled and incubated for 30-60 min.
Materials:
- EbbaBiolight
- LB broth (w/o salt)
- Bacteria on standard culture plate
- Sterile glass coverslips (24x24 mm)
- 6-well plate with cover or adhesive seal
- Mounting medium
- Nail polish
- EtOH 70%
Equipment:
- Incubator (28°C)
- Shaking Incubator (37°C)
- Fluorescence microscope
Assay Procedure:
- Plate preparation:
- place two sterile glass coverslips opposite to each other and inclined towards the walls of the wells in a 6-well plate.
-
Prepare bacterial inoculum:
- Pick a colony from a standard culture plate.
- Transfer colony to LB broth.
- Prepare an overnight or exponential culture under continuous shaking at 37°C.
- Dilute bacterial culture 1:100 in fresh LB broth.
- Add EbbaBiolight (1:1000) and mix gently.
- Pipett 6 ml of EbbaBiolight supplemented bacterial culture into each well fitted with glass coverslips.
- Incubation:
- Seal the plate with cover or adhesive seal & incubate at a suitable temperature for 24-48 h.
- Mounting & Sealing:
- Remove the glass coverslips from each well & wipe the backside.
- Add a drop (ca 30 µl) of mounting medium to the sample and flip the coverslip on a microscope slide.
- Remove excess liquid, seal the coverslip with nail polish & clean the slide with 70% EtOH.
- Endpoint analysis:
- Visualize biofilm using fluorescence microscopy. Use filter-sets or excitation- and emission parameters as indicated in the table below.
Read more →



